Difference between revisions of "Tracer's gellary: C. elegans case study"
Shahar.maoz (Talk | contribs) (New page: We have used the Tracer to visualize and explore different execution traces of the biological system model described in Kam, N., Harel, D., Kugler, H., Marelly, R., Pnueli, A., Hubbard...) |
Shahar.maoz (Talk | contribs) m (C. elegans moved to Tracer's gellary: C. elegans case study) |
||
| (2 intermediate revisions by one user not shown) | |||
| Line 1: | Line 1: | ||
We have used [[the Tracer]] to visualize and explore different execution traces of the biological system model described in Kam, N., Harel, D., Kugler, H., Marelly, R., Pnueli, A., Hubbard, E.J.A., Stern, M.J.: Formal Modeling of C. elegans Development: A Scenario-Based Approach (C. Priami (ed.) Proc. 1st Int. Workshop on Computational Methods in Systems Biology (CMSB'03), Lecture Notes in Computer Science, vol. 2602, pp. 4-20. Springer (2003)). The model deals with the process of vulval precursor cell fate determination in the development of the C. elegans nematode worm. The model was implemented in the Play-Engine tool. It consists of more than 400 scenarios, divided into 22 use cases. Typical traces are 300-400 events long, and involve 40-80 different scenarios from the model. | We have used [[the Tracer]] to visualize and explore different execution traces of the biological system model described in Kam, N., Harel, D., Kugler, H., Marelly, R., Pnueli, A., Hubbard, E.J.A., Stern, M.J.: Formal Modeling of C. elegans Development: A Scenario-Based Approach (C. Priami (ed.) Proc. 1st Int. Workshop on Computational Methods in Systems Biology (CMSB'03), Lecture Notes in Computer Science, vol. 2602, pp. 4-20. Springer (2003)). The model deals with the process of vulval precursor cell fate determination in the development of the C. elegans nematode worm. The model was implemented in the Play-Engine tool. It consists of more than 400 scenarios, divided into 22 use cases. Typical traces are 300-400 events long, and involve 40-80 different scenarios from the model. | ||
| − | Due to the large number of scenarios in the C. elegans model, using the various horizontal | + | Due to the large number of scenarios in the C. elegans model, using the various horizontal filters proved crucial. Moreover, typical traces included several periods with large numbers of multiple active instances of the same scenario (up to 30 such). The Multiplicities supporting view helped us in exploring the details of these multiple copies. |
| − | We examined a number of execution traces of this model. Differences between the traces were due to the probabilistic choices inside the LSCs in the model, as played-out by the Play-Engine, and due to our use of | + | We examined a number of execution traces of this model. Differences between the traces were due to the probabilistic choices inside the LSCs in the model, as played-out by the Play-Engine, and due to our use of different initial configurations, simulating experiments with the worm's wild-type and the various mutations and cell ablations that appear in the C. elegans literature. Using the trace comparison features, we were able to find where such different `experiments' show different behavior and where their behaviors are equivalent. |
| − | We thank Itai Segall for implementing trace generation in the Play-Engine and for his help with using this model. | + | We thank Itai Segall for implementing trace generation in the Play-Engine and for his help with using this model. |
Main view and overview: | Main view and overview: | ||
Latest revision as of 13:32, 18 October 2010
We have used the Tracer to visualize and explore different execution traces of the biological system model described in Kam, N., Harel, D., Kugler, H., Marelly, R., Pnueli, A., Hubbard, E.J.A., Stern, M.J.: Formal Modeling of C. elegans Development: A Scenario-Based Approach (C. Priami (ed.) Proc. 1st Int. Workshop on Computational Methods in Systems Biology (CMSB'03), Lecture Notes in Computer Science, vol. 2602, pp. 4-20. Springer (2003)). The model deals with the process of vulval precursor cell fate determination in the development of the C. elegans nematode worm. The model was implemented in the Play-Engine tool. It consists of more than 400 scenarios, divided into 22 use cases. Typical traces are 300-400 events long, and involve 40-80 different scenarios from the model.
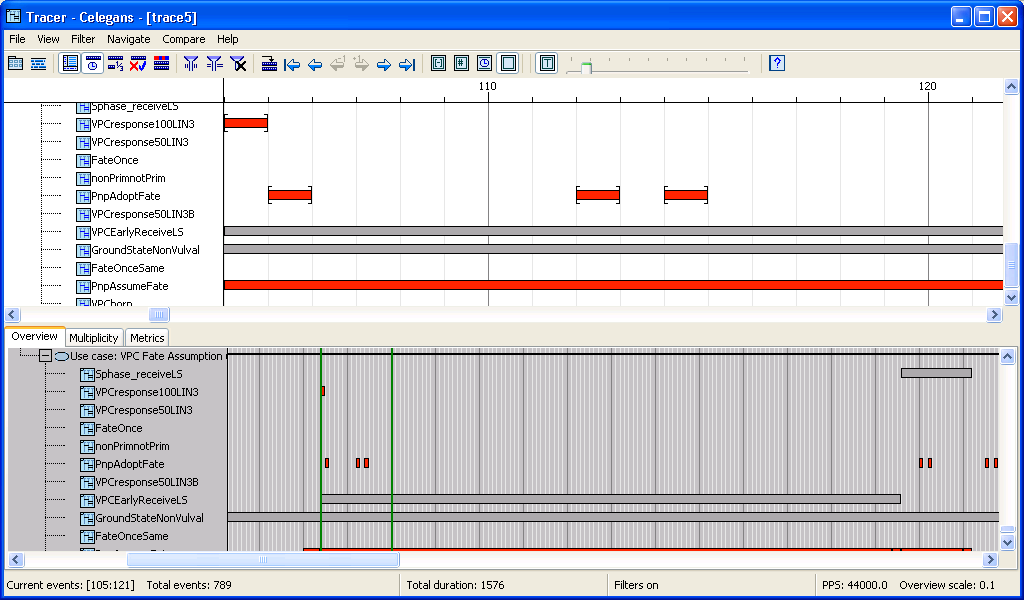
Due to the large number of scenarios in the C. elegans model, using the various horizontal filters proved crucial. Moreover, typical traces included several periods with large numbers of multiple active instances of the same scenario (up to 30 such). The Multiplicities supporting view helped us in exploring the details of these multiple copies.
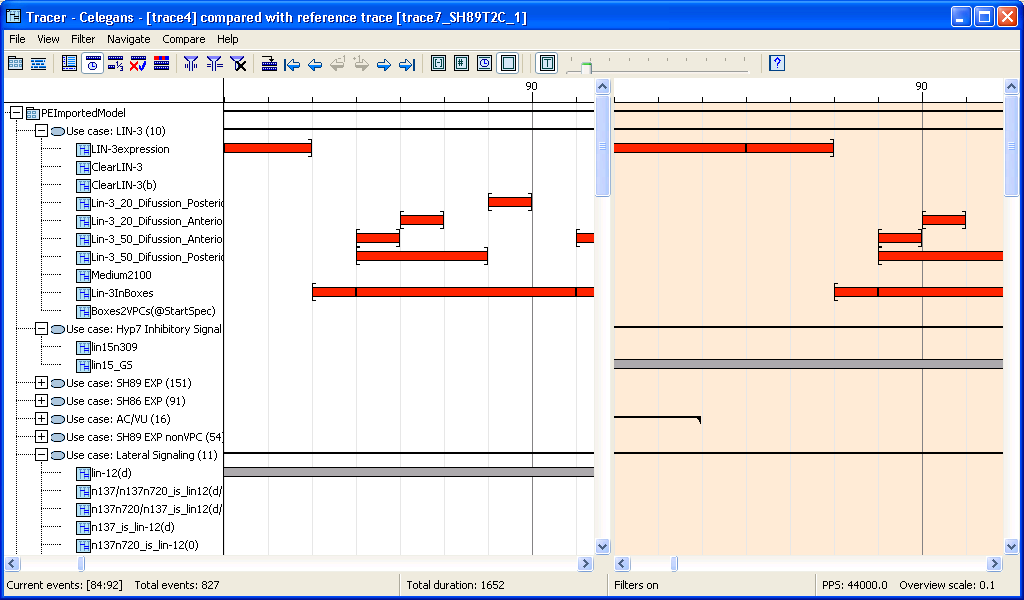
We examined a number of execution traces of this model. Differences between the traces were due to the probabilistic choices inside the LSCs in the model, as played-out by the Play-Engine, and due to our use of different initial configurations, simulating experiments with the worm's wild-type and the various mutations and cell ablations that appear in the C. elegans literature. Using the trace comparison features, we were able to find where such different `experiments' show different behavior and where their behaviors are equivalent.
We thank Itai Segall for implementing trace generation in the Play-Engine and for his help with using this model.
Main view and overview:
Comparing traces: